EHR-Based Genomic Discovery
As part of the Electronic Medical Records and Genomics (eMERGE) Network, the research team in Dr. Kullo's Atherosclerosis and Lipid Genomics Laboratory is conducting electronic health record (EHR)-based genetic studies.
The network is funded by the National Human Genome Research Institute (NHGRI) to develop and implement approaches for leveraging biorepositories with EHR systems for large-scale genomic research, including genome-wide association studies (GWAS), sequencing and structural variation.
The five initial eMERGE sites were:
- Group Health Cooperative (with the University of Washington)
- Marshfield Clinic
- Mayo Clinic
- Northwestern University
- Vanderbilt University
Related publications
Mayo eMERGE phase I project
The Mayo Clinic eMERGE I project aimed to identify genetic loci associated with peripheral artery disease and red blood cell traits, including hemoglobin, hematocrit, red blood cell count, mean corpuscular volume, mean corpuscular hemoglobin and mean corpuscular hemoglobin concentration.
Phenotype characteristics and covariates relevant to statistical genetic analyses were derived using EHR-based algorithms, including diagnosis and procedure codes, laboratory data, medication use, and natural language processing of unstructured text. An ethical, legal and social issues (ELSI) component was included and focused in particular on community interaction and return of results.
Mayo eMERGE phase II project
In phase II of eMERGE (July 2011 to June 2015), the research infrastructure established in eMERGE-I was leveraged to identify common genetic variants that influence medically important phenotypes.
The Mayo eMERGE-II cohort (6,916 people) includes the 3,769 eMERGE-I patients and an additional 3,147 people, the majority (90 percent) of whom were genotyped on the same Illumina 660W platform. A major focus in phase II was to translate recent GWAS findings into clinical practice.
Related publications
- Jarvik GP, Amendola LM, Berg JS, Brothers K, Clayton EW, Chung W, Evans BJ, Evans JP, Fullerton SM, Gallego CJ, Garrison NA, Gray SW, Holm IA, Kullo IJ, Lehmann LS, McCarty C, Prows CA, Rehm HL, Sharp RR, Salama J, Sanderson S, Van Driest SL, Williams MS, Wolf SM, Wolf WA; eMERGE Act-ROR Committee and CERC Committee; CSER Act-ROR Working Group, Burke W. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet. 2014;94(6):818-26. [PMID: 24814192; PMC4121476].
- Kullo IJ, Haddad R, Prows CA, Holm I, Sanderson SC, Garrison NA, Sharp RR, Smith ME, Kuivaniemi H, Bottinger EP, Connolly JJ, Keating BJ, McCarty CA, Williams MS, Jarvik GP. Return of results in the genomic medicine projects of the eMERGE network. Front Genet. 2014;5:50. [PMID: 24723935; PMC3972474].
- Hazin R, Brothers KB, Malin BA, Koenig BA, Sanderson SC, Rothstein MA, Williams MS, Clayton EW, Kullo IJ. Ethical, legal, and social implications of incorporating genomic information into electronic health records. Genet Med. 2013;15(10):810-6. [PMID: 24030434; PMC3926430].
Mayo eMERGE phase III project

The eMERGE III study is an exciting collaboration between Mayo Clinic and Mountain Park Health Center in Arizona that's funded by the NHGRI and the Mayo Clinic Center for Individualized Medicine.
Mayo Clinic's eMERGE III study investigated the return of genetic test results relevant to hypercholesterolemia and colon polyps.
As part of the study, genetic testing for hypercholesterolemia and colon polyps will be performed at a National Institutes of Health laboratory. A team of expert Mayo Clinic investigators will look for the presence of genetic abnormalities causing hypercholesterolemia or colon polyps.
The genetic results will be entered into the EHR. Patients will receive genetic counseling and support from Mayo Clinic physicians and genetic counselors.
In eMERGE III, the participating sites were expanded and include:
- Children's Hospital of Philadelphia
- Cincinnati Children's Hospital Medical Center
- Columbia University
- Geisinger Health System
- Group Health Research Institute/University of Washington
- Mayo Clinic
- Northwestern University
- Seattle Brigham and Women's Hospital
- Vanderbilt University School of Medicine
Study 1: EHR-based algorithms to ascertain vascular disease phenotypes
With the decreasing cost of genotyping and the increasing availability of genomic data from consortia, selection of phenotypes of interest becomes a critical part of phenotype-genotype association studies.
As an important node of the eMERGE Network, our Atherosclerosis and Lipid Genomics Laboratory designed several EHR-based phenotyping algorithms that can be found on the Phenotype KnowledgeBase (PheKB) website. PheKB is a collaborative environment enabling cross-site collaboration for algorithm development, validation and sharing for reuse with confidence.
Related publications
- Liu H, Bielinski SJ, Sohn S, Murphy S, Wagholikar KB, Jonnalagadda SR, Ravikumar KE, Wu ST, Kullo IJ, Chute CG. An information extraction framework for cohort identification using electronic health records. AMIA Jt Summits Transl Sci Proc. 2013 March 18, 149-53. [PMID: 24303255; PMC3845757].
- Sohn S, Ye Z, Liu H, Chute CG, Kullo IJ. Identifying abdominal aortic aneurysm cases and controls using natural language processing of radiology reports. AMIA Summit on Clinical Research Informatics Proc. 2013 March 13, 249-253. [PMID: 24303276; PMC3845740].
- Savova GK, Fan J, Ye Z, Murphy SP, Zheng J, Chute CG, Kullo IJ. Discovering peripheral arterial disease cases from radiology notes using natural language processing. AMIA Annu Symp Proc. 2010 Nov 13, 722-6. [PMID: 21347073; PMC3041293].
- Kullo IJ, Ye Z. ePhenotyping for abdominal aortic aneurysm in the eMERGE Network: algorithm development and KNIME workflow. (JAMA, in review).
- Safarova MS, Liu H, Kullo IJ. Rapid identification of familial hypercholesterolemia from electronic health records. Poster presentation at the American Heart Association Scientific Sessions, Nov. 9, 2015, in Orlando, Florida.
- Ding K, de Andrade M, Manolio TA, Crawford DC, Rasmussen-Torvik LJ, Ritchie MD, Denny JC, Masys DR, Jouni H, Pachecho JA, Kho AN, Roden DM, Chisholm R, Kullo IJ. Genetic variants that confer resistance to malaria are associated with red blood cell traits in African-Americans: an electronic medical record-based genome-wide association study. G3 (Bethesda). 2013 Jul 8;3(7):1061-8. [PMID: 23696099; PMC3704235].
- Ding K, Shameer K, Jouni H, Masys DR, Jarvik GP, Kho AN, Ritchie MD, McCarty CA, Chute CG, Manolio TA, Kullo IJ. Genetic Loci implicated in erythroid differentiation and cell cycle regulation are associated with red blood cell traits. Mayo Clin Proc. 2012;87(5):461-74. [PMID: 22560525; PMC3538470].
- Kullo IJ, Ding K, Jouni H, Smith CY, Chute CG. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS One. 2010;5(9):e13011. [PMID: 20927387; PMC2946914].
Study 2: Genome-wide association studies
We have performed EHR-based genome-wide association studies of peripheral artery disease and red blood cell indices.
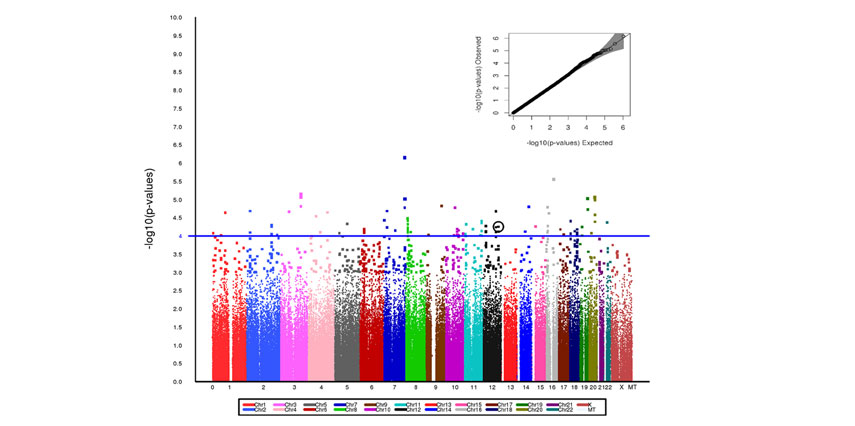
A Manhattan plot representative of the genome-wide association for peripheral artery diseases cases and controls

Manhattan and quantile-quantile plots for genome-wide association analysis of erythrocyte sedimentation rate in the eMERGE cohort
Related publications
- Ding K, de Andrade M, Manolio TA, Crawford DC, Rasmussen-Torvik LJ, Ritchie MD, Denny JC, Masys DR, Jouni H, Pachecho JA, Kho AN, Roden DM, Chisholm R, Kullo IJ. Genetic variants that confer resistance to malaria are associated with red blood cell traits in African-Americans: an electronic medical record-based genome-wide association study. G3 (Bethesda). 2013 Jul 8;3(7):1061-8. [PMID: 23696099; PMC3704235].
- Ding K, Shameer K, Jouni H, Masys DR, Jarvik GP, Kho AN, Ritchie MD, McCarty CA, Chute CG, Manolio TA, Kullo IJ. Genetic Loci implicated in erythroid differentiation and cell cycle regulation are associated with red blood cell traits. Mayo Clin Proc. 2012;87(5):461-74. [PMID: 22560525; PMC3538470].
- Kullo IJ, Ding K, Jouni H, Smith CY, Chute CG. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS One. 2010;5(9):e13011. [PMID: 20927387; PMC2946914].
Study 3: Phenome-wide association studies (PheWAS)
PheWAS is a unique tool to look at traits associated with a particular therapy, enabling discovery of pleiotropic effects of genes.
This strategy mines the full potential of the EHR for genome-phenome associations, rapid and efficient validation of known associations between genes of interest, and interindividual variation in traits.
Our Atherosclerosis and Lipid Genomics Lab previously detected signals of natural selection in PCSK9, and recent animal and genetic studies implicating PCSK9 in the regulation of blood pressure and triglyceride-rich lipoprotein metabolism indicate that genetic variants in PCSK9 may have pleiotropic effects.
We are conducting a comprehensive investigation of pleiotropic effects of PCSK9.

A Manhattan plots summarizing results of phenome-wide association analyses of (A) platelet count and (B) mean platelet volume. Results are from logistic regression analysis for 1,368 EMR-defined phenotypes in 13,688 individuals, adjusted for age and sex.
Related publications
- Cronin RM, Field JR, Bradford Y, Shaffer CM, Carroll RJ, Mosley JD, Bastarache L, Edwards TL, Hebbring SJ, Lin S, Hindorff LA, Crane PK, Pendergrass SA, Ritchie MD, Crawford DC, Pathak J, Bielinski SJ, Carrell DS, Crosslin DR, Ledbetter DH, Carey DJ, Tromp G, Williams MS, Larson EB, Jarvik GP, Peissig PL, Brilliant MH, McCarty CA, Chute CG, Kullo IJ, Bottinger E, Chisholm R, Smith ME, Roden DM, Denny JC. Phenome-wide association studies demonstrating pleiotropy of genetic variants within FTO with and without adjustment for body mass index. Front Genet. 2014;5:250. [PMID: 25177340; PMC4134007].
- Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, Field JR, Pulley JM, Ramirez AH, Bowton E, Basford MA, Carrell DS, Peissig PL, Kho AN, Pacheco JA, Rasmussen LV, Crosslin DR, Crane PK, Pathak J, Bielinski SJ, Pendergrass SA, Xu H, Hindorff LA, Li R, Manolio TA, Chute CG, Chisholm RL, Larson EB, Jarvik GP, Brilliant MH, McCarty CA, Kullo IJ, Haines JL, Crawford DC, Masys DR, Roden DM. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102-10. [PMID: 24270849; PMC3969265].
- Shameer K, Denny JC, Ding K, Jouni H, Crosslin DR, de Andrade M, Chute CG, Peissig P, Pacheco JA, Li R, Bastarache L, Kho AN, Ritchie MD, Masys DR, Chisholm RL, Larson EB, McCarty CA, Roden DM, Jarvik GP, Kullo IJ. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet. 2014;133(1):95-109. [PMID:24026423; PMC3880605].
- Simonti CN, Vernot B, Bastarache L, Bottinger E, Carrell DS, Chisholm RL, Crosslin DR, Hebbring SJ, Jarvik GP, Kullo IJ, Li R, Pathak J, Ritchie MD, Roden DM, Verma SS, Tromp G, Prato JD, Bush WS, Akey JM, Denny JC, Capra JA. The phenotypic legacy of admixture between modern humans and Neandertals. Science. 2016 Feb 12;351(6274):737-41. doi: 10.1126/science.aad2149. [PMID:26912863].